 |
 |
 |
 |
 |
 |
 |
Peroxisome biogenesis, function, and dynamics
Bonnie Bartel, Professor
Department of Biosciences
bartel@rice.edu
publications
Peroxisomes are eukaryotic organelles that are essential for life in plants and animals. Peroxisomes sequester various oxidative reactions, thereby enhancing metabolic efficiency while protecting cytosolic constituents from oxidative damage. Although our knowledge of peroxisome function and dysfunction is increasing, mechanistic understanding of how this critical organelle is formed, maintained, and recycled remains incomplete.
We are seeking to fill these knowledge gaps by tackling the following questions: How and where do peroxisomes originate? How and why do intralumenal vesicles form within peroxisomes? How are proteins imported into the organelle? How do peroxisomes interact with other organelles? How is peroxisomal quality control enforced?
We address these questions using Arabidopsis thaliana; the unique peroxisomal functions, small size, and facile genetics of this model allow straightforward modulation of peroxisomal processes in an intact multicellular organism. Moreover, the relatively large size of plant peroxisomes (compared to yeast and mammalian peroxisomes) offers exceptional opportunities to decipher peroxisome biogenesis and membrane intricacies using live-cell imaging.
Our peroxisome research articles
Our pexophagy publications
Our IBA publications
Our peroxisome-related review articles
We are grateful for current and prior support for this research from the NIH (R35GM130338, R01GM079177), the NSF (MCB-1516966, MCB-0745122, IBN-0315596), and the Robert A. Welch Foundation (C-1309).
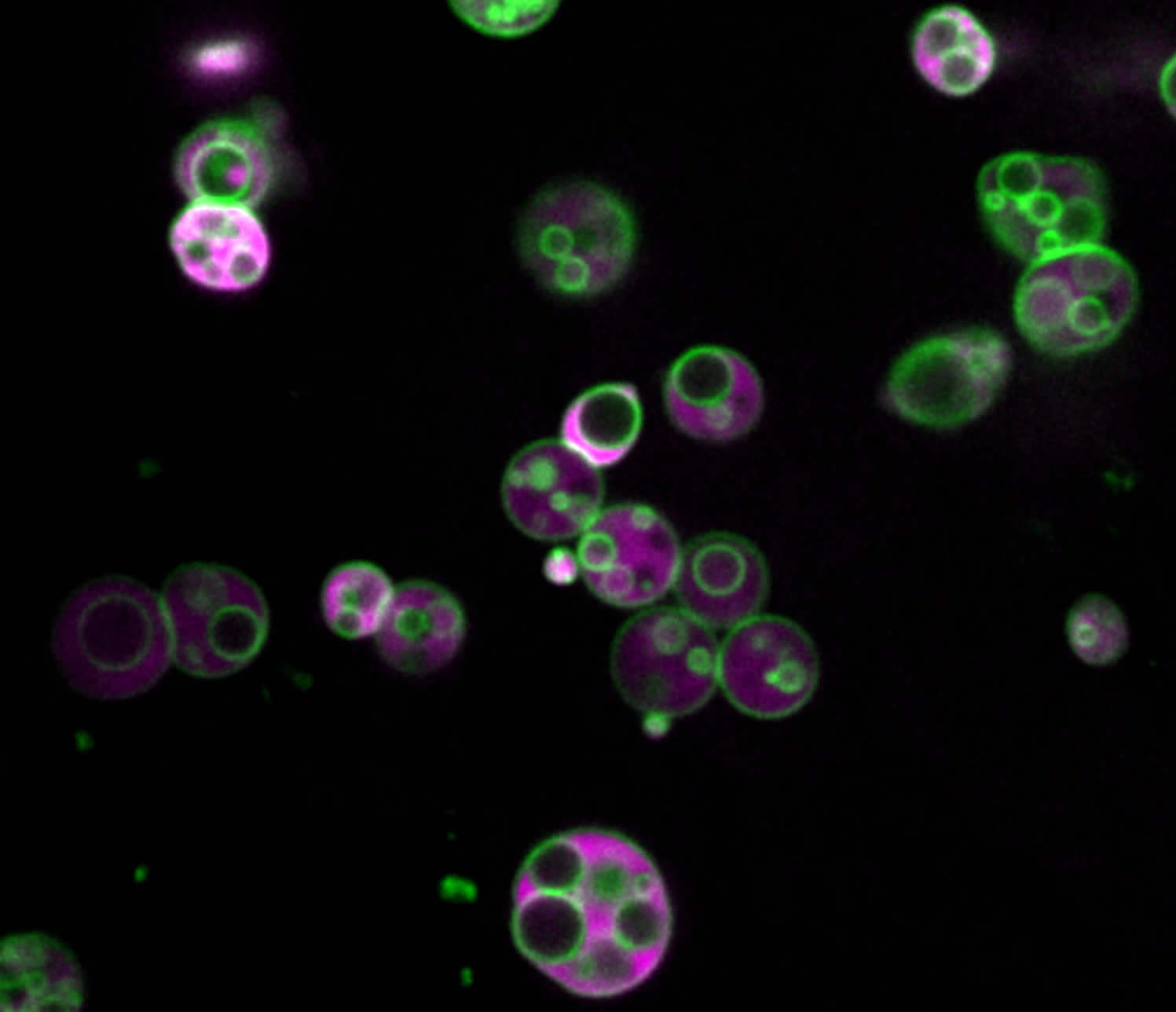
Peroxisomes contain intralumenal vesicles. Peroxisomes in an Arabidopsis seedling expressing reporter marking peroxisome membrane (green) and lumen (magenta). Image credit: Zachary Wright.
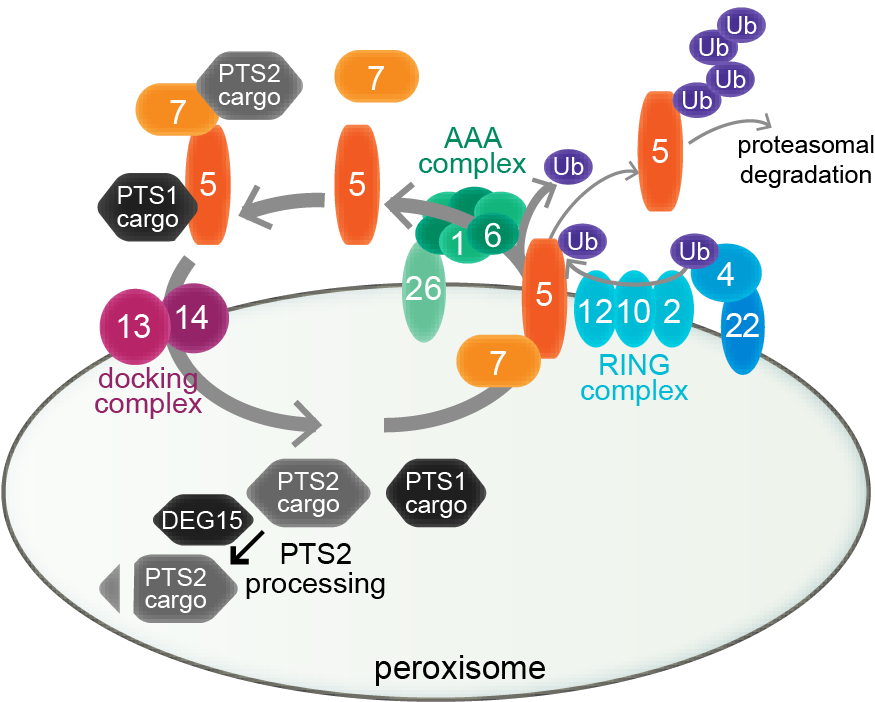
Peroxisome biogenesis. Peroxisome biogenesis employs a core set of conserved peroxins (numbered ovals) to import lumenal proteins, which are synthesized in the cytosol and targeted to the peroxisome via PEX5 interaction with a C-terminal PTS1 (black proteins) or PEX7 interaction with an N-terminal PTS2 (gray proteins). PEX5 docks with a PEX13-PEX14 complex for cargo delivery; PEX7 binding to PEX5 (in plants and mammals) is required for PTS2-protein delivery. Receptor-recycling peroxins remove PEX5 via mono-ubiquitination by the PEX4 Ub-conjugating enzyme and the PEX12 RING peroxin, allowing removal by the PEX1-PEX6 ATPase complex. PEX5 is poly-ubiquitinated and degraded by the proteasome when recycling is slowed. DEG15 clips the PTS2 signal after matrix protein import.
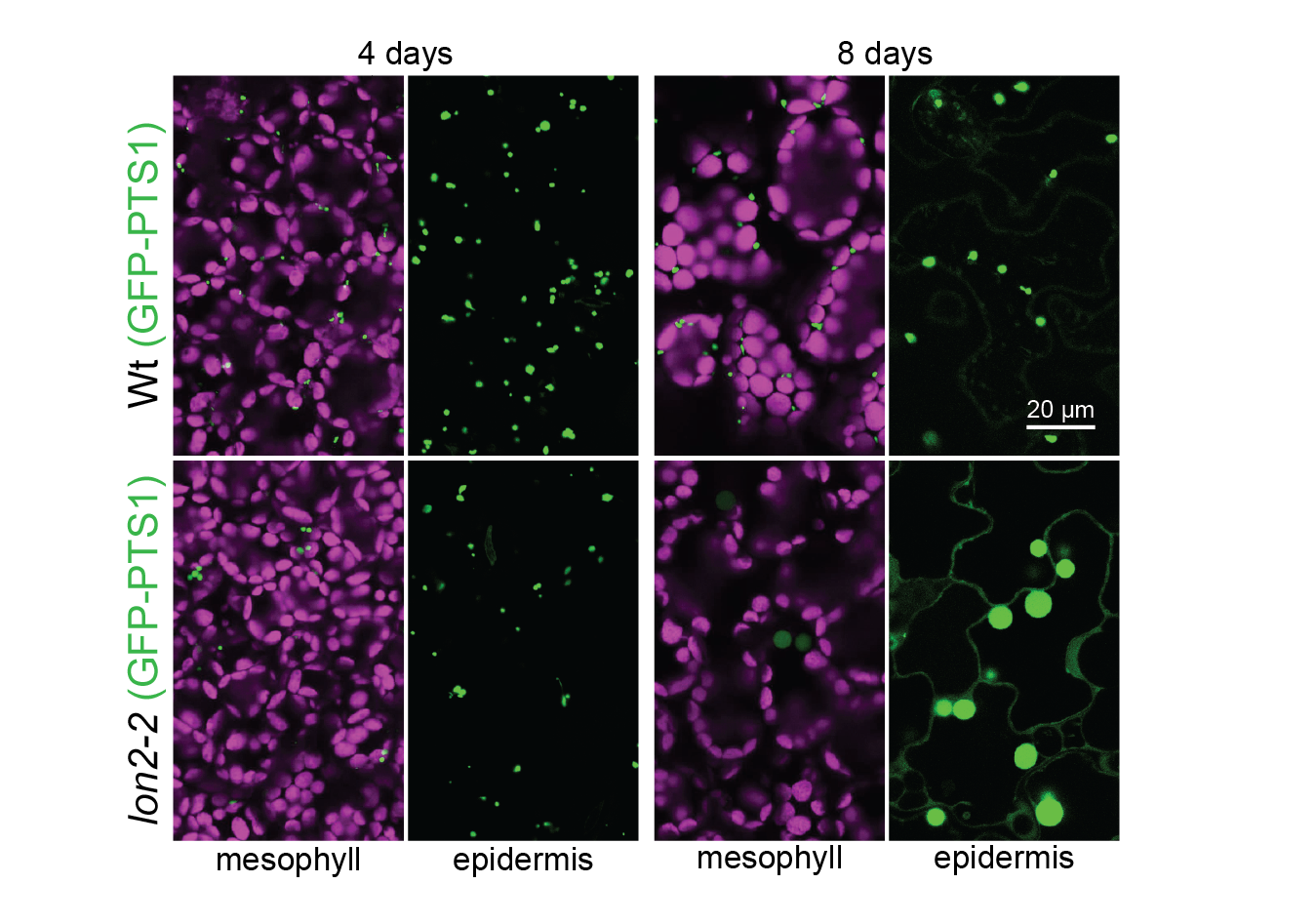
Autophagy of peroxisomes. When the LON2 peroxisomal protease is dysfunctional, peroxisomes are degraded via autophagy and GFP-PTS1 (green) that is peroxisomal in young seedlings becomes cytosolic as seedlings age. Chlorophyll autofluorescence is in magenta. Image credit: Mauro Rinaldi.